Basics
Table of contents
Obtaining a MGX Project
MGX Roles
Metadata
Connecting to Server
Uploading own files
Importing reference genomes
Search
Obtaining a MGX project
Currently, there is no way to automatically create new MGX projects. If you would like to analyze your metagenome or metatranscriptome data with MGX, please send a mail to the MGX team, which can be contacted at:
MGX projects and users are managed by the General Project Management System (GPMS) developed at CeBiTec (Bielefeld University), which provides Single Sign-On (SSO) for all our applications.
To apply for a new project, please provide:
- a short project name, e.g. MGX_AcidMine
- a one-line description of your project (Acid Mine Drainage metagenome)
- a contact address of a single person responsible for the project (PI or group leader) and the corresponding GPMS login name.
MGX Roles
MGX offers different access levels (roles), which are assigned individually for each project:
Adminsare equal toUsers, but can request access to be granted to additional users.Usershave full access to a MGX project and are able to define new or modify present datasets, import new sequences and execute analysis jobs. They are also able to delete all data associated with a project.Guestsare provided read-only access to a project, i.e. they are able to access all information already present, view analysis results and export data; however, they are unable to perform new analysis or delete data from the project.
For all MGX projects, the person requesting the project is automatically added as an Admin. As new users can always be added to an existing MGX project, all registered users are required to carefully protect their login credentials and not to share them with any third party.
Metadata
In addition to the sequence data, MGX requires a user to provide additional information about a dataset, e.g. further details about the investigated habitat as well as sampling and sequencing procedures. Metadata in the MGX platform is organized in a hierarchical manner describing
- the geographical location of a habitat
- the sample taken from a habitat
- the DNA extraction procedure
- sequencing technology and protocol
Connecting to a MGX server
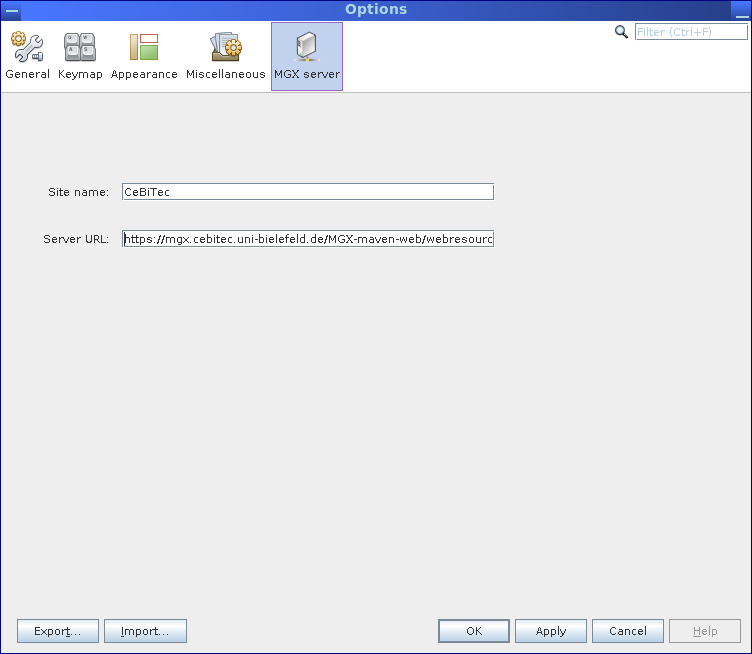
After installation, the MGX application is already preconfigured to connect to the MGX server instance hosted at Justus Liebig University Giessen (JLU). In case a different MGX server should be used, the default server can be changed choosing Tools → Options from the menu and navigating to the MGX server tab. While the site name can be freely chosen by the user, the server URL has to be entered as provided by the site administrators.
A different default server instance can optionally be configured in the MGX server tab, which is available from the Tools →Options menu.
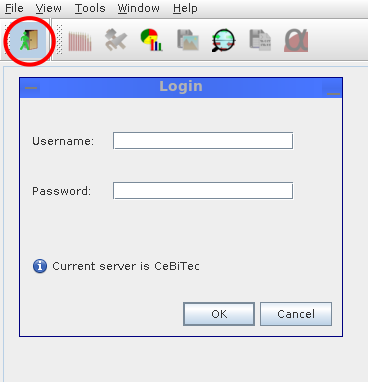
The first button in the menu toolbar will bring up the login dialog, allowing to connect to the configured server; the login screen also reflects the name of the current server.
All communication between the MGX user interface and the MGX server is encrypted using the standardized SSL (Secure Sockets Layer) protocol, ensuring confidentiality of unpublished data and protecting the integrity of login credentials.
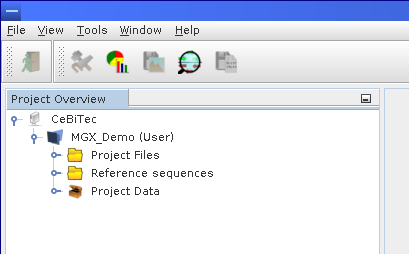
After successfully logging in, the Project Overview window lists all available projects a user is allowed to access, including both public as well as private projects. For each project, the role of the current user is given in brackets. Projects are easily opened or closed by simply expanding the corresponding nodes in the Project Explorer window.
After successful login, the Project Explorer component will automatically open, showing a list of MGX projects available to the current user. Shown is just one project, MGX_Demo, with User access level indicated behind the project name.
Divided into four different sections, a MGX project offers (from top to bottom) dedicated storage for files to be used by analysis pipelines, managed reference sequences (including annotation data, if available) and general project data containing metadata as well as sequence datasets. An additional section provides access to metagenome and metatranscriptome assemblies.
Each project contains metagenome datasets as well as structured storage, where user-provided databases can be uploaded to be used in custom analysis pipelines.
Uploading own files
For each project, MGX provides dedicated storage to allow users to provide custom data, which can subsequently be used with analysis pipelines. Thus, own sequence collections or hidden Markov models HMM model files for genes of interest can easily be uploaded and later included in analysis pipelines.
Each project includes flat storage where custom data can be stored, e.g., own FASTA files to be used as reference databases for metagenome analysis.
While it may be necessary to implement your own pipeline depending on the desired kind of analysis, the MGX repository hosts predefined pipeline templates addressing the most common cases.
The BestHit-Blast template can be used to annotate metagenome sequences with the description of a Blast hit after the user has uploaded a FASTA file containing amino acid sequences, and the BestHit-HMM template provides the same functionality for HMMs.
Importing reference genomes
MGX provides several analysis pipelines to align metagenome reads to reference genomes. Before these pipelines can be used, the corresponding reference genome has to be added to the project. There are two possible ways to achieve this:
- MGX repository hosts published reference sequences
- Annotated reference genomes obtained from the NCBI
iIn addition, users may choose to upload a custom reference sequence in FASTA, GenBank, or EMBL format, e.g., a finished but unpublished genome not available from official sources.
Import: Reference sequences for mapping targets may be imported from the global repository or uploaded in EMBL/GenBank/FASTA format.
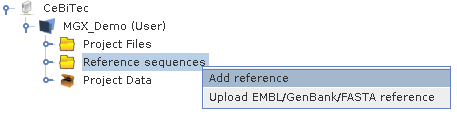
To add a reference genome: right-click on the Reference sequences node within the project view and select either Add reference to access the MGX repository or Upload EMBL/GenBank/FASTA reference to provide your own sequence.
Once the import is complete, the reference genome is available for analysis and can be selected for the corresponding analysis pipelines that provide reference mapping, e.g., BowTie or FR-HIT.
Search
Search: Icon for the metagenome Search component.
Example
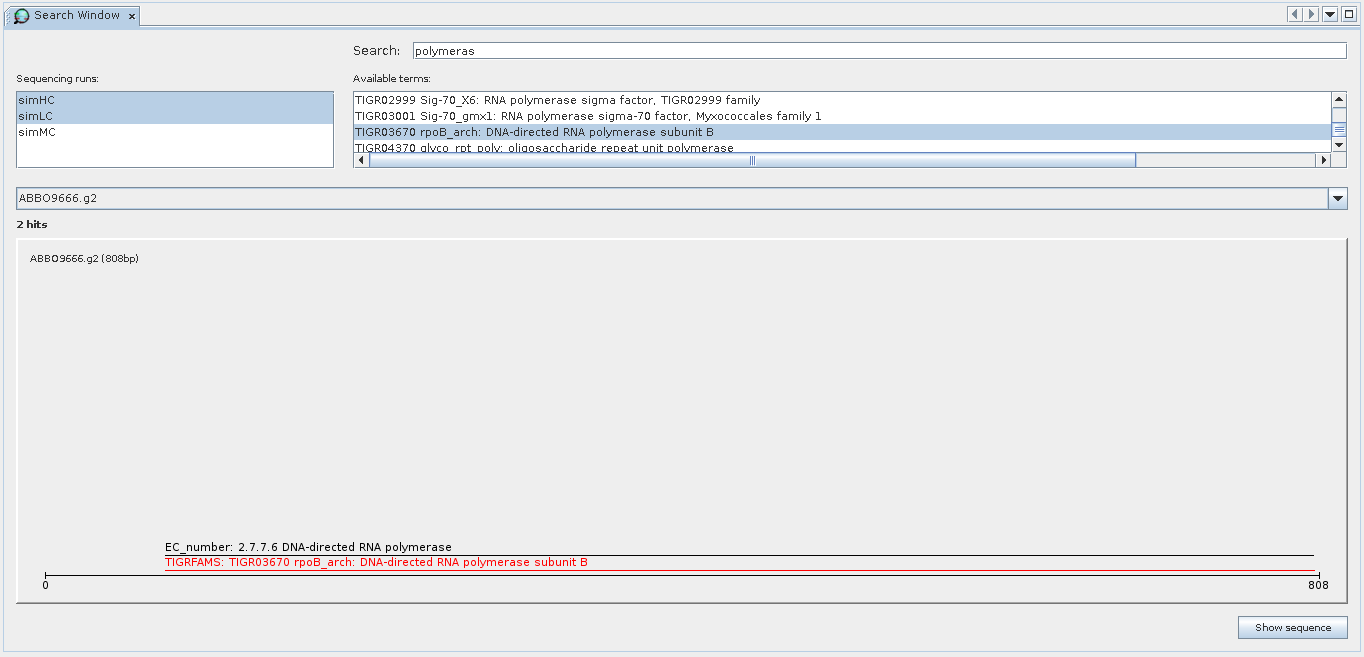
Search component showing results for the term polymeras. The search was performed within the select metagenomes simHC and simLC (top left).
The bottom part shows an individual sequence identified by the search. Search results are displayed together with all other attributes available for sequences, thus allowing the identification of the co-occurrence of results.